Regression Diagnostics for Linear Models
diagnostics.lm.Rd# S3 method for lm
diagnostics(x, alpha = 0.4, span = 0.8, plot = TRUE, ...)Arguments
- x
an object of class
"lm"- alpha
numeric; transparency for plot points (default=0.4)
- span
numeric; smoothing parameter for loess fit lines (default=0.8)
- plot
logical; If
TRUE(the default), graphs are printed. Otherwise, they are returned invisibly.- ...
not currently used
Value
A five component list containing ggplot2 graphs:
qqplot, crplots, slplot, vifplot, and influenceplot.
Details
The diagnostics function is a wrapper for several
diagnostic plotting functions:
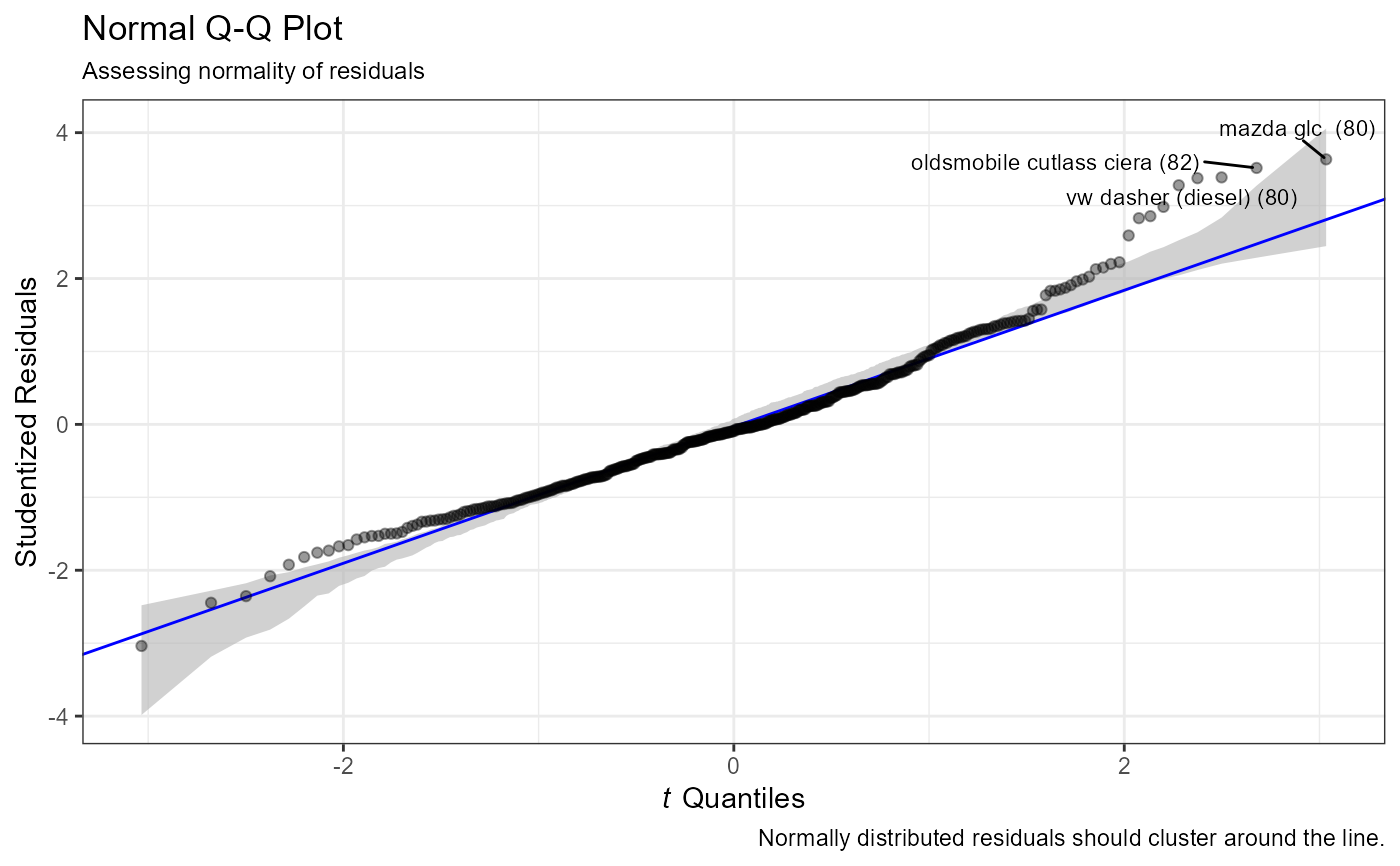
- Normality
Normality of the (studentized) residuals is assessed via a Normal Q-Q plot (
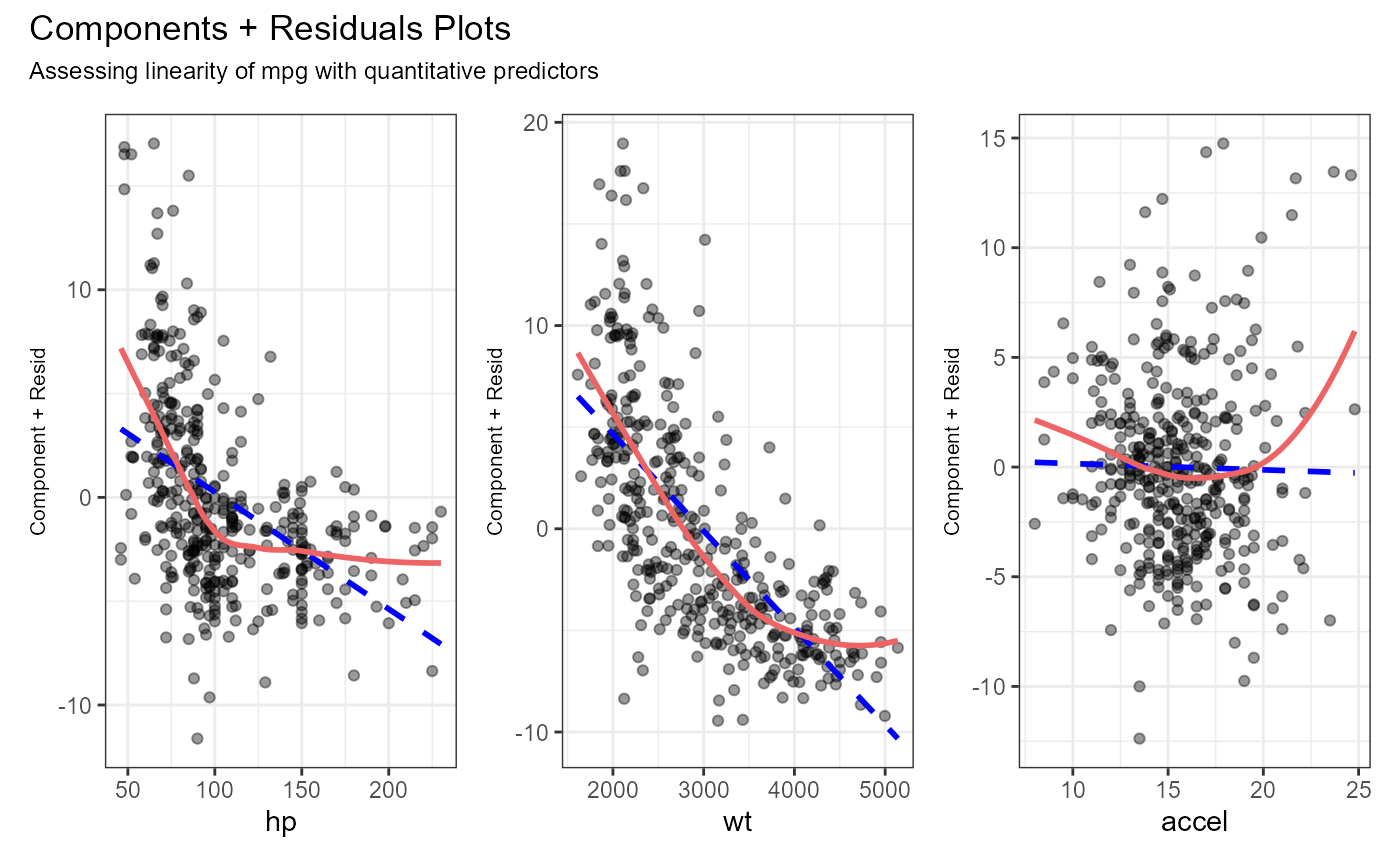
qq_plot).- Linearity
Linearity of the explanatory-response relationships are assessed via Component + Residual (partial residual) plots (
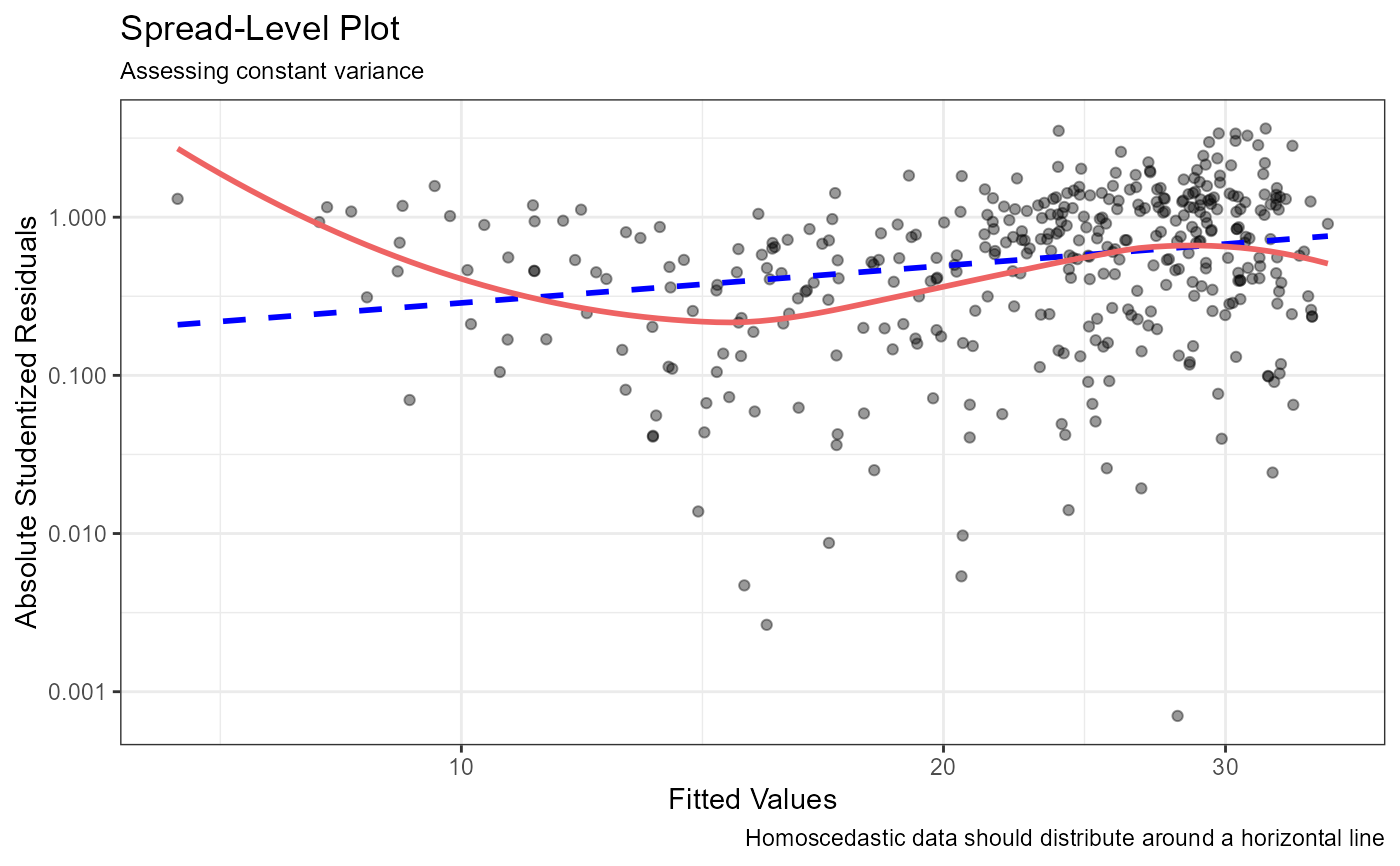
cr_plots). If there is a single predictor, a scatter plot with linear and loess lines is produced.- Constant variance
Homoscedasticity is evaluated via a Spread-Level plot (
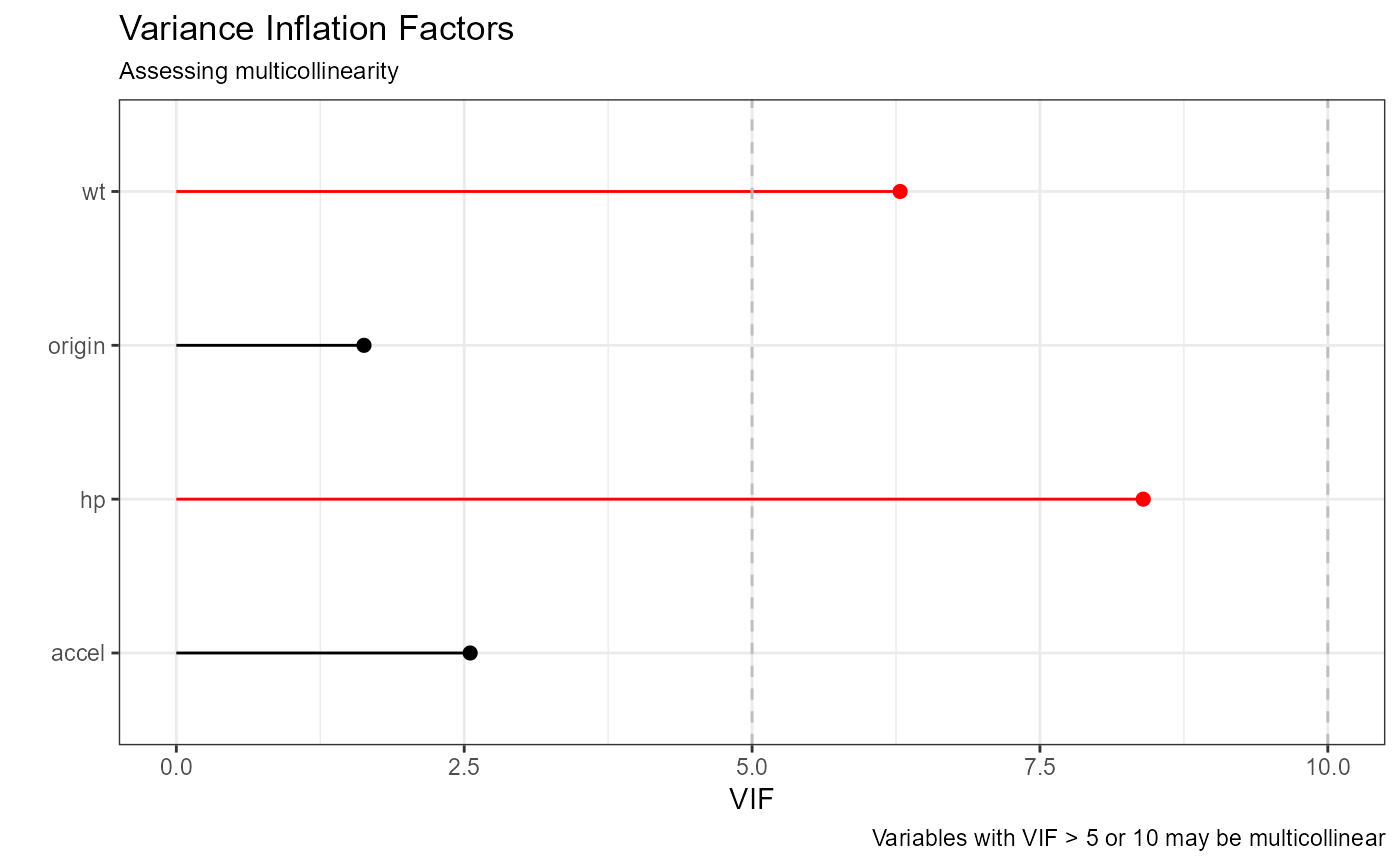
spread_plot).- Multicollinearity
Multicollinearity is assessed via variance inflation factors (
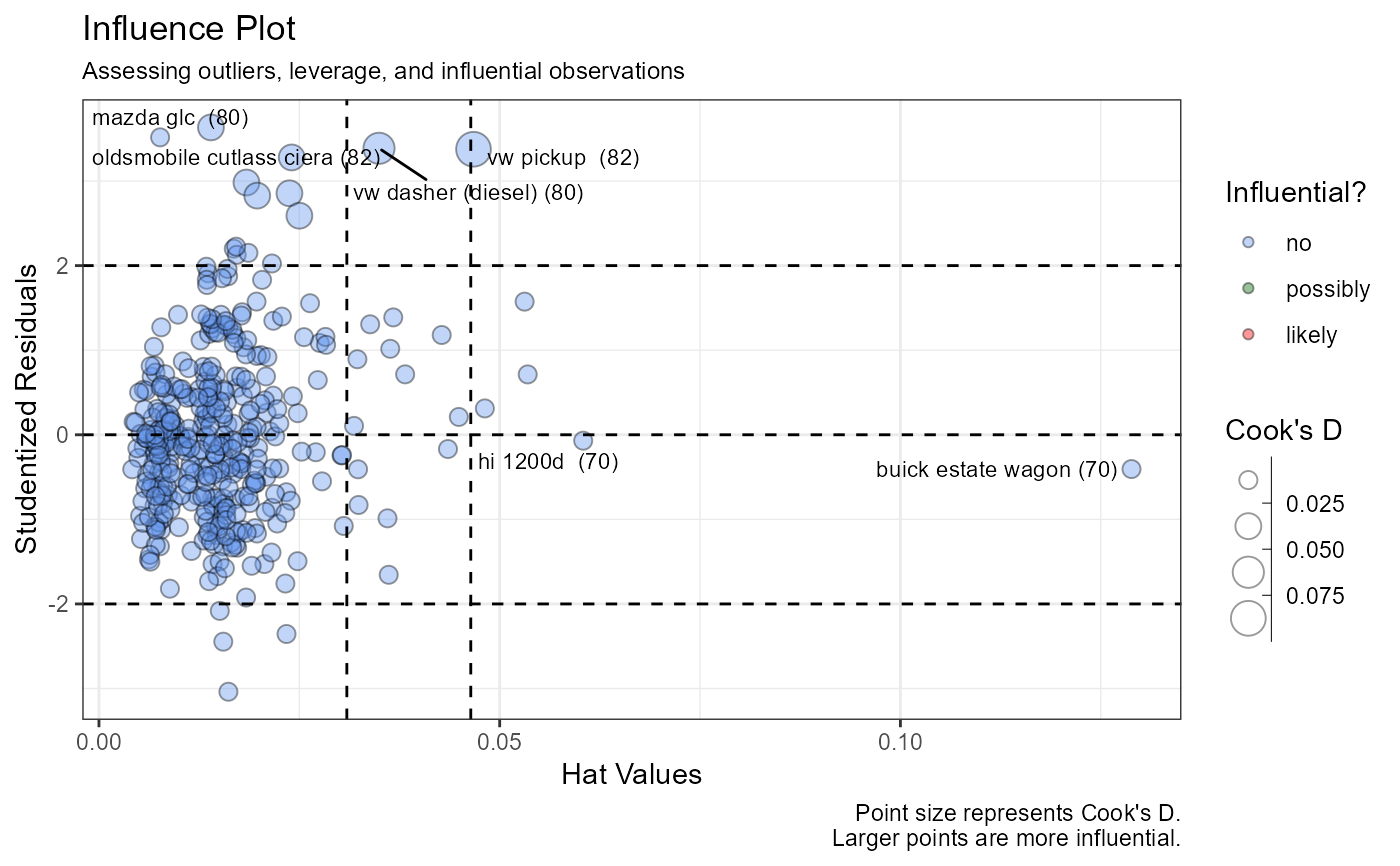
vif_plot). If there is a single predictor variable, this section is skipped.- Outliers, leverage, and influence
A influence plot identifies outliers and influential observations (
influence_plot).
Note
Each function relies heavily on the car package. See the
help for individual functions for details.
See also
Examples
fit <- lm(mpg ~ hp + wt + accel + origin, data = auto_mpg)
diagnostics(fit)